Cell Tree Rings
Smarter and cheaper way to measure scalable single cell somatic mutation burden across the life sciences market space
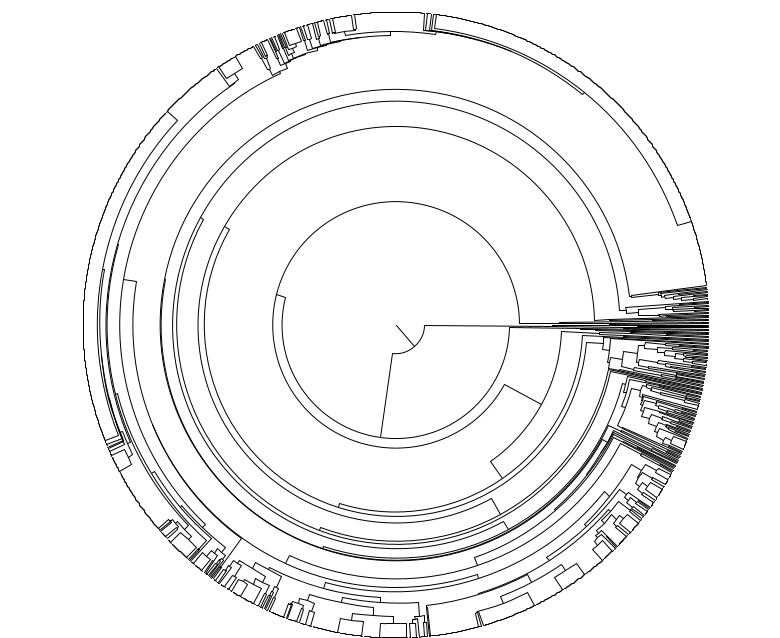
Smarter and cheaper way to measure scalable single cell somatic mutation burden across the life sciences market space

Cell Tree Rings is the first industry-developed, scalable and application focused tool that is ready to unleash the power of evolutionary cell lineage trees using somatic mutations from thousands of individual cells.
Current aging clocks used to assess healthy longevity interventions do not measure somatic mutation burden, which informs of the foundational aging hallmark genome instability. Not measuring existing somatic mutation burden before an intervention, the current default practice, can potentially offset biological age assessment by decades, and not assessing the potential changes in that burden due to and after the intervention can miss important additional mechanisms, effects and opportunities. We believe that Cell Tree Rings can serve as a core component for combined aging clocks to mitigate current uncertainty in the assessment of geroprotective trials.
Currently we are working with the Healthy Longevity Clinic to turn the initial results into clinically useful and actionable insights by analysing the actual underlying somatic mutations in the accessible human genes.
The applications of Cell Tree Rings are very broad and the tool can be directly used in clonal hematopoiesis of indeterminate potential (CHIP) studies, drug efficacy testing and cell therapies.
Earlier write-up on Cell Tree Rings in the industry insider Longevity Technology called Using somatic evolution as a human aging timer.
Please see our proof-of-principle study on biorxiv: Cell Tree Rings: the structure of somatic evolution as a human aging timer
View study
